Variation in Mutation Rates
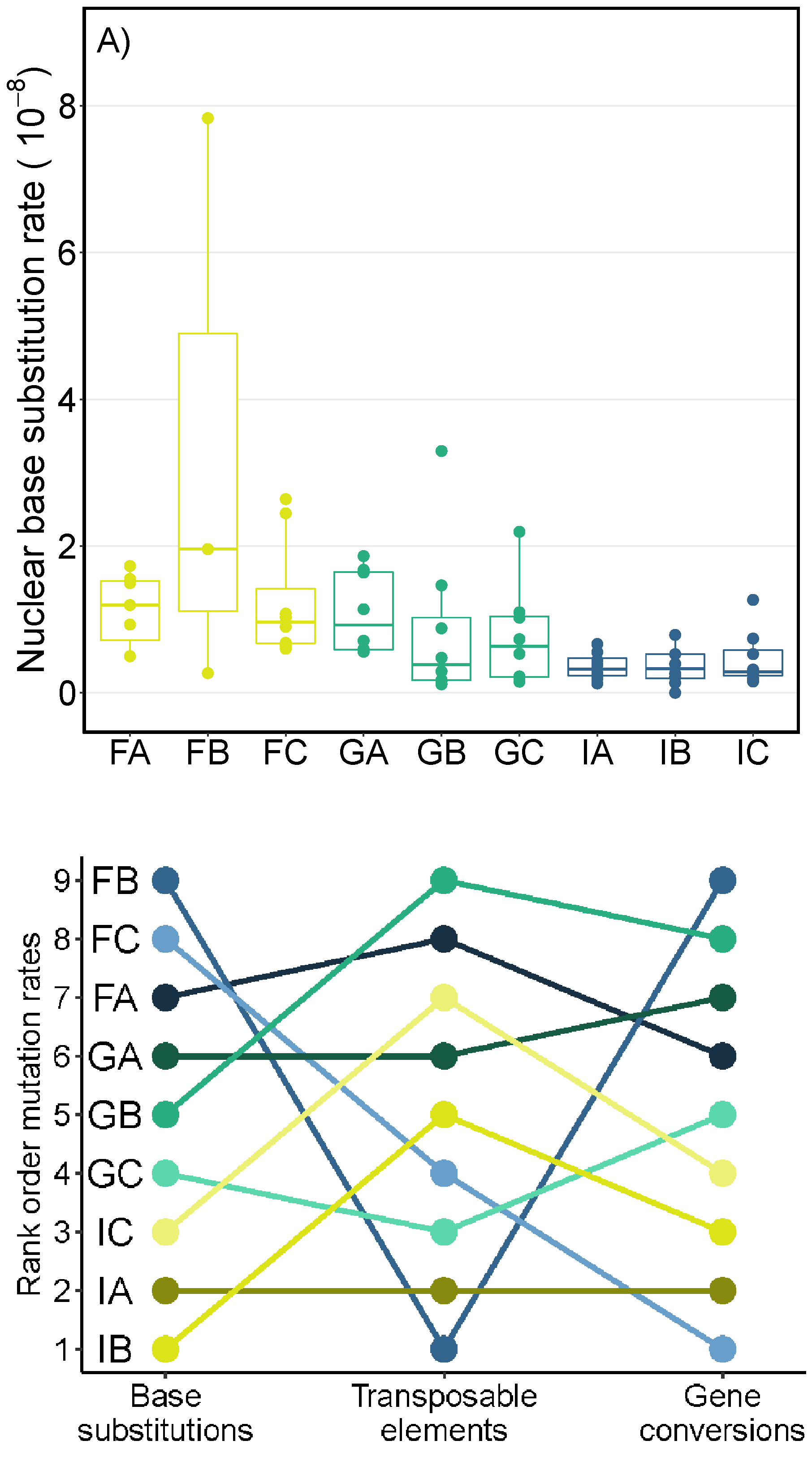
Mutations are the ultimate source of genetic variation. Although it has become easier to obtain good estimate of mutation rates, we still know very little about how these rates vary within and between species. My research utilizes bioinformatic analyses to estimate the mutations rates for different types of mutations (SNPs, indels, CNVs, microsatellites, transposable elements) from mutation accumulation lines of Daphnia magna. I have found significant intraspecific variation in mutations rates between genotypes and populations as well as between the nuclear and mitochondrial genome (Ho et al. 2020). It is also becoming clear that rates for different mutation types do not strongly covary across genotypes (Ho et al. 2019). These results have implications for the evolution of mutation rates and their impacts on the genome.
Genetic Consequences of Uniparental Reproduction
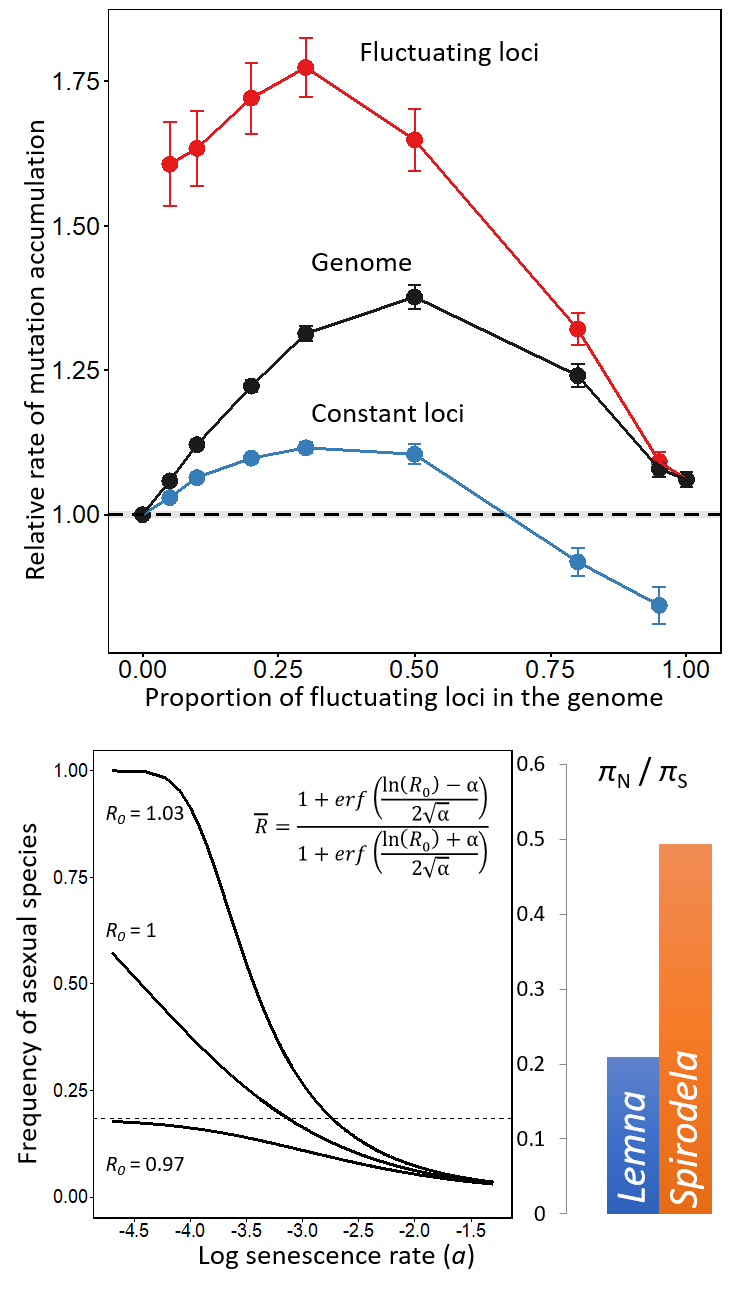
Despite the advantages of asexual reproduction, the majority of species are sexual/outcrossing. The rarity of highly asexual/selfing species may be attributed to their low efficacy of selection, leading to the accumulation of deleterious mutations and eventual extinction. By incorporating biologically realistic parameters to population genetics theory, I showed that: i) the rate of mutation accumulation is elevated in selfing species experiencing fluctuations in selection (Ho et al. 2018), ii) asexual/selfing species are less likely to outcompete sexual/outcrossing species when mutation loads are inherited (Ho et al. 2017). In addition to my theoretical work, I have also identified genomic signatures of asexual reproduction and low selection efficacy in the duckweed, Spirodela polyrhiza (Ho et al. 2019).